3 Week4 - Algorithms & Approximations
3.1 Lesson 1 - Algorithms
3.1.1 Shortest Path Problem
Relations <- read.table(
text = "
From To weight
SL CH 300
SL IN 245
SL LV 263
SL NV 312
CH CL 362
CH IN 201
IN CO 176
IN CI 112
IN LV 114
LV LX 86
LV NV 175
CL CO 142
CL HB 322
CL MT 201
CL CN 251
CO CI 105
CI LX 95
CI CN 204
LX CN 177
LX KV 170
KV GR 299
CN MT 157
CN GR 244
CN RI 318
MT HB 213
MT WA 209
GR RI 205
HB WA 120
WA RI 111
", header =T)
net <- graph_from_data_frame(Relations, directed=F)
#TODO Erro no pacote
# plot(net,vertex.label.cex = 0.8 , vertex.color=Relations$From, edge.label=Relations$weight,
# edge.label.cex = 0.8, edge.color = "gray70", edge.label.color = "black")
#
#
# as_edgelist(net, names=T)
# as_adjacency_matrix(net, attr="weight")
#
# as_data_frame(net)
#Plot the degree distribution for our network:
deg <- degree(net, mode="all")
deg.dist <- degree_distribution(net, cumulative=T, mode="all")
plot(x=0:max(deg), y=1-deg.dist, pch=19, cex=1.2, col="orange",
xlab="Degree", ylab="Cumulative Frequency")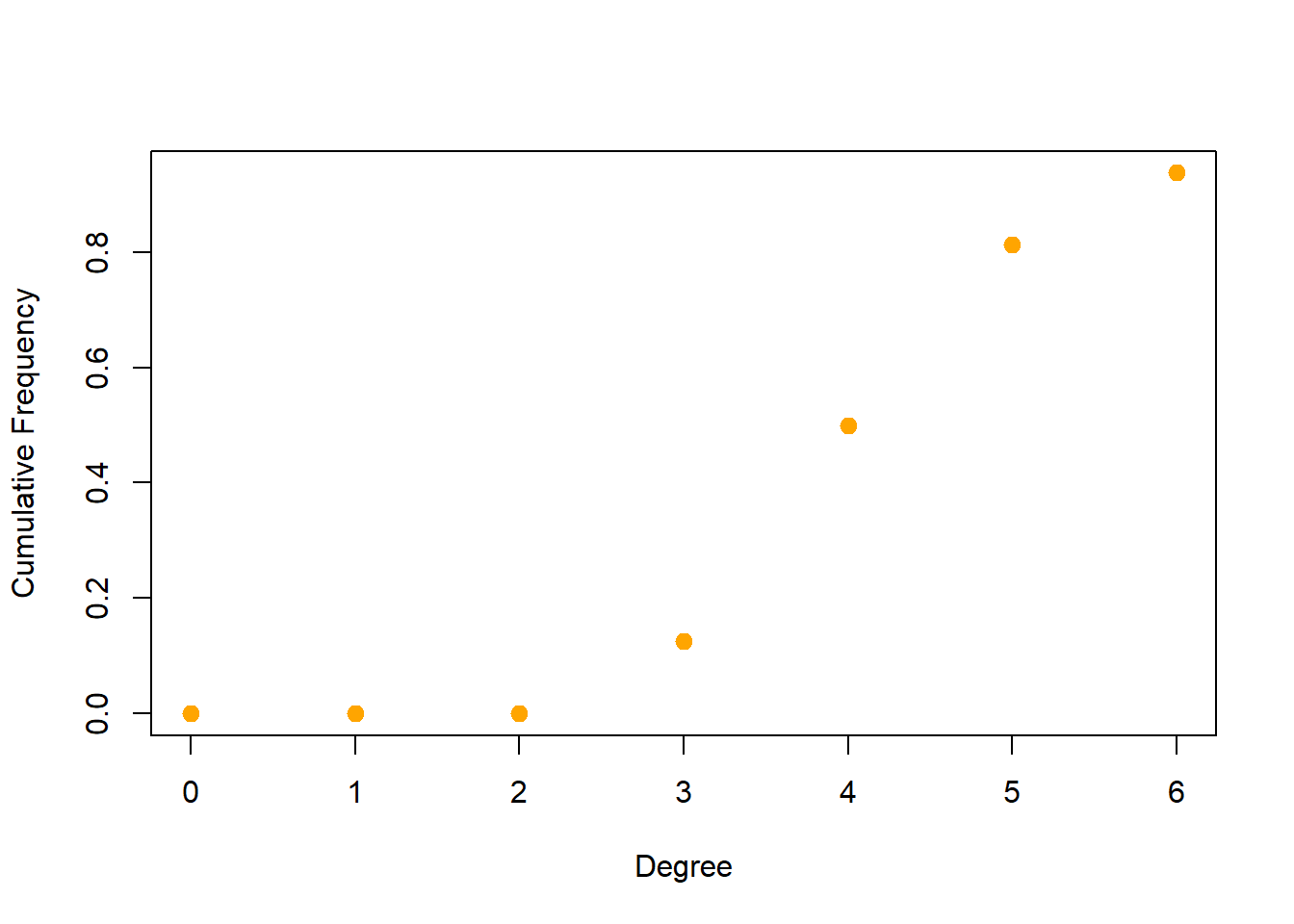
#Shortest Path
sPath <- function(net, from, to){
#Function created based on https://kateto.net/tutorials/
Path <- shortest_paths(net, from = from, to = to, weights = E(net)$weight, output = "both")
# Generate edge color variable to plot the path:
ecol <- rep("gray80", ecount(net))
ecol[unlist(Path$epath)] <- "orange"
# Generate edge width variable to plot the path:
ew <- rep(2, ecount(net))
ew[unlist(Path$epath)] <- 4
#Generate node color variable to plot the path:
vcol <- rep("gray40", vcount(net))
vcol[unlist(Path$vpath)] <- "gold"
#plot
plot(net, vertex.color=vcol, edge.color=ecol,
edge.width=ew, edge.arrow.mode=0)
}
sPath(net, from = "LV", to = "WA")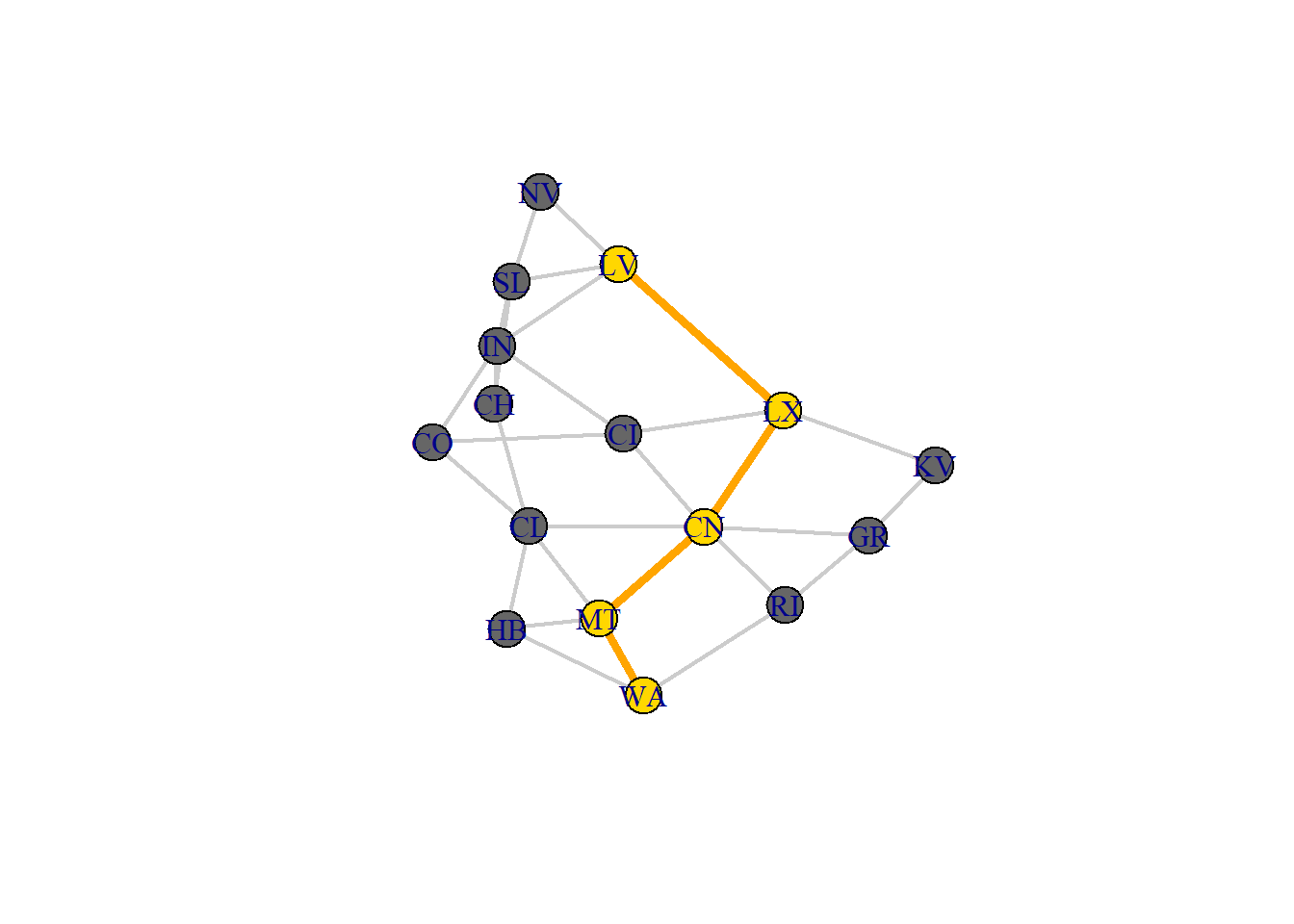
All lowest distances from the model
# all vertices to all vertices
distMatrix <- shortest.paths(net, v=V(net), to=V(net), weight = net$weight)
kable(distMatrix, caption = "Lowest distance from node to node") %>%
kable_styling() %>%
scroll_box(width = "100%")| SL | CH | IN | LV | CL | CO | CI | LX | KV | CN | MT | GR | HB | WA | NV | RI | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SL | 0 | 300 | 245 | 263 | 563 | 421 | 357 | 349 | 519 | 526 | 683 | 770 | 885 | 892 | 312 | 844 |
| CH | 300 | 0 | 201 | 315 | 362 | 377 | 313 | 401 | 571 | 517 | 563 | 761 | 684 | 772 | 490 | 835 |
| IN | 245 | 201 | 0 | 114 | 318 | 176 | 112 | 200 | 370 | 316 | 473 | 560 | 640 | 682 | 289 | 634 |
| LV | 263 | 315 | 114 | 0 | 428 | 286 | 181 | 86 | 256 | 263 | 420 | 507 | 633 | 629 | 175 | 581 |
| CL | 563 | 362 | 318 | 428 | 0 | 142 | 247 | 342 | 512 | 251 | 201 | 495 | 322 | 410 | 603 | 521 |
| CO | 421 | 377 | 176 | 286 | 142 | 0 | 105 | 200 | 370 | 309 | 343 | 553 | 464 | 552 | 461 | 627 |
| CI | 357 | 313 | 112 | 181 | 247 | 105 | 0 | 95 | 265 | 204 | 361 | 448 | 569 | 570 | 356 | 522 |
| LX | 349 | 401 | 200 | 86 | 342 | 200 | 95 | 0 | 170 | 177 | 334 | 421 | 547 | 543 | 261 | 495 |
| KV | 519 | 571 | 370 | 256 | 512 | 370 | 265 | 170 | 0 | 347 | 504 | 299 | 717 | 615 | 431 | 504 |
| CN | 526 | 517 | 316 | 263 | 251 | 309 | 204 | 177 | 347 | 0 | 157 | 244 | 370 | 366 | 438 | 318 |
| MT | 683 | 563 | 473 | 420 | 201 | 343 | 361 | 334 | 504 | 157 | 0 | 401 | 213 | 209 | 595 | 320 |
| GR | 770 | 761 | 560 | 507 | 495 | 553 | 448 | 421 | 299 | 244 | 401 | 0 | 436 | 316 | 682 | 205 |
| HB | 885 | 684 | 640 | 633 | 322 | 464 | 569 | 547 | 717 | 370 | 213 | 436 | 0 | 120 | 808 | 231 |
| WA | 892 | 772 | 682 | 629 | 410 | 552 | 570 | 543 | 615 | 366 | 209 | 316 | 120 | 0 | 804 | 111 |
| NV | 312 | 490 | 289 | 175 | 603 | 461 | 356 | 261 | 431 | 438 | 595 | 682 | 808 | 804 | 0 | 756 |
| RI | 844 | 835 | 634 | 581 | 521 | 627 | 522 | 495 | 504 | 318 | 320 | 205 | 231 | 111 | 756 | 0 |
3.1.1.1 Dijkstra’s Algorithm
Inputs: n Connected graph with nodes and arcs with positive costs, d(ij) n Source (s) and Terminal (t) nodes
Algorithm:
for all nodes in graph, set L()=∞, P()=Null, S()=0
set s to i, S(i)=1, and L(i)=0
For all nodes, j, directly connected (adjacent) to node i, if L(j) > L(i) + d(ij), then set L(j) = L(i) + d(ij) and P(j)=i
For all nodes where S()=0, select the node with lowest L() and set it to i, set S(i)=1
Is this node t, the terminal node? If so, go to end If not, go to step 3
end – return L(t)
The shortest_path and shortest.path function already solves it by default with dijkstra’s model when the weights have only positive values.
But let’s mimic the algorithm and try to get the results.
Djikstraz <- function(net, source, terminal){
source <- 1
terminal <- 7
data <- as.matrix(as_adjacency_matrix(net, attr="weight"))
n = nrow(data)
M = 10000000
iMin = 1
data <- cbind(data, L = M)
data <- cbind(data, P = 0)
data <- cbind(data, S = 0)
data[source, "S"] = 1
data[source, "L"] = 0
i = source
while (i != terminal) {
print(i,terminal)
for (j in 1:n) {
print(paste0("j = ",j))
if (data[j, "L"] > 0) {
if (data[j, "L"] == data[i, "L"] + data[i, j]) {
data[j, "L"] <- data[i, "L"] + data[i, j]
data[j, "P"] <- i
}
}
}
Min <- M
for (i in 1:n) {
if (data[i, "S"] == 0) {
if (data[i, "L"] < Min) {
Min <- data[i, "L"]
iMin <- i
}
}
}
i <- iMin
data[i, "S"] = 1
}
data
}3.1.1.2 LP
$$ \[\begin{align*} \textbf{Minimize} & \ z = \sum_{i=1}^{N} \sum_{j=1}^{M} c_{i,j} \cdot x_{i,j} \\ \textbf{subject to} \\ & \sum_{i=1}^{N} \sum_{j=1}^{M} x_{i,j} - x_{j,i} = 0, \ \forall j \neq s, \ j \neq t \\ & \sum_{i=1}^{N} x_{i,j} = 1, \ \forall j = t \\ & \sum_{i=1}^{N} x_{j,i} = 1 \ \forall j = s \\ & x_{i} \ge 0 \\ \textbf{Where} \\ & x_{i,j} = \text{Number of units flowing on nodes} \\ & c_{i,j} = \text{Cost per unit for flow} \\ & s = \text{Source node} \\ & t = \text{Terminal node} \end{align*}\] $$
3.1.3 Vehicle Routing Problem
3.1.3.2 Clark-Wright
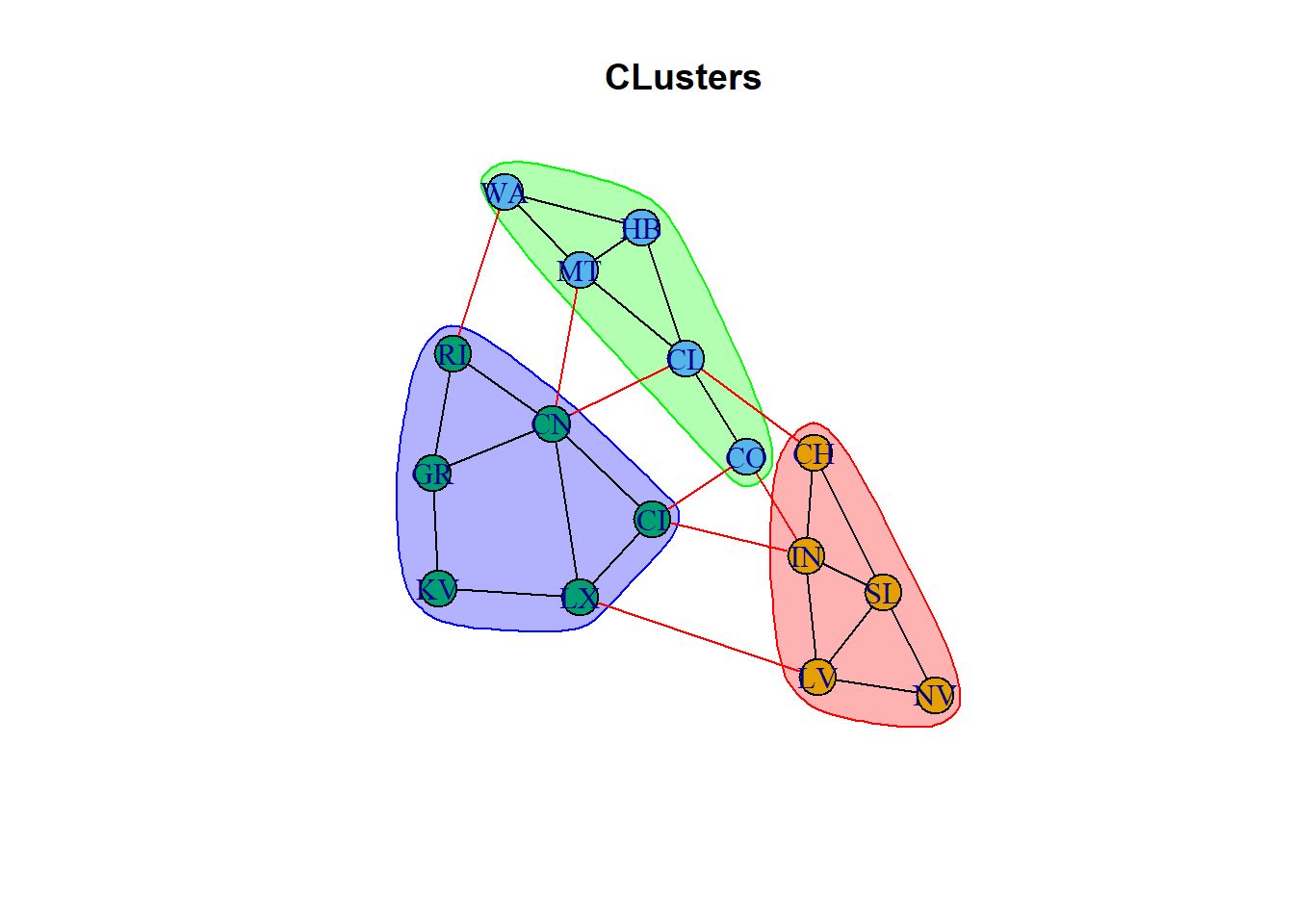
Cool way to create the clusters based on the distance weights, clustering based on the lowest interconnected nodes distance. This is cool because we don’t have to define the number of clusters, it will define it by the distance parameter
# edge betweenness
ceb <- cluster_edge_betweenness(net, weights = Relations$weight)
#Dendogram
dendPlot(ceb, mode="hclust")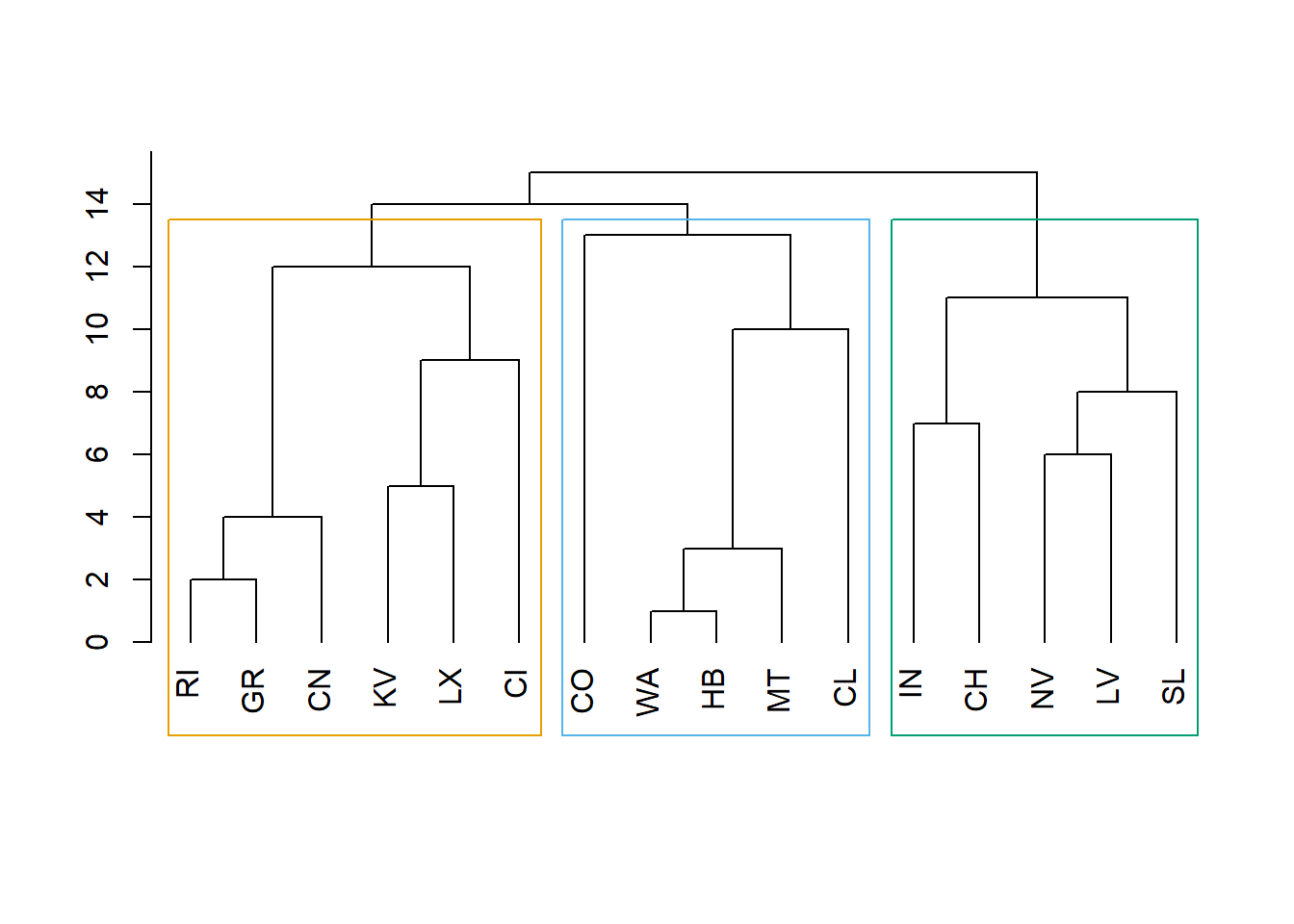
#Graph Plot
plot(ceb, net, main = "CLusters")